Wow it's been a long time since I updated this blog. There's a very good reason for that-- I had all these great plans about how I was going to update as all my results came in and give a real-time look at the progress of science. Well, I have done exactly that, because the progress of science has been next to nil, especially in the lab. Here's the status of my Funaria project as of this week:
Phenotypes: For 300 sporophytes of Funaria, I took pictures of the seta (stalk), the capsule, and the spores, and tallied various things about them. I then used various statistical tests to observe some things about my Funaria populations, with some interesting results. For example, I found that within F. hygrometrica, there is significant correlation between size and latitude- sporophytes furthest north (like Ithaca, NY and Brattleboro, VT) were much smaller than sporophytes furthest south (like Durham, NC and Catlett, VA). This latitude gradient would be the perfect thing to explore further with DNA approaches...
Lab Work: I've now spent three months trying to develop microsatellites for Funaria, to use as DNA fingerprinting and paternity tests. Using two different methods, I've come up with only four sites in the whole genome that have the small repeating regions, and I don't even know if those are variable enough to use. This presents a problem, as microsatellites are expensive to develop and I don't really have much funding. Despite the promising results from my phenotype work, it appears I have a decision to make about the future of my PhD work.
Meanwhile, this semester I have adopted the project left over from another graduate student, for a class that I am taking. Originally it was just going to be me tying together some loose ends on the project and using the class as an excuse to take care of it. The project involves peatmoss, or Sphagnum, specifically the two species that I helped collect in my very first experience with mosses. I still have pictures from that field trip up here. It is a group of species that has always interested me, but I didn't want to "step on the toes" of another grad student's work. Now that he is no longer with the lab, further exploration of this group could fall to me. But should I switch projects now, after a year dead set on Funaria? I have to weigh the pros and cons of each:
Funaria hygrometrica
Advantages: Weedy, easy to find, grows in discrete clumps for easy population sampling, grows well in the greenhouse, has a short generation time, bisexual gametophytes.
Disadvantages: Expensive genetic marker development has not gone well, strong possibility of uninteresting results, between species issues unclear.
Sphagnum macrophyllum
Advantages: Extremely well developed genetic markers, known populations, interesting questions already established, project funded by the lab. Between-species issues easily studied.
Disadvantages: Does not grow well in the greenhouse, crossing experiments unlikely, populations much more spread out, many are asexual.
Part of the difficulty of deciding between them is that their advantages seem complimentary-- where one is unfit, the other thrives! I struggled with the decision for a month, but some signs kept popping up: I kept failing to identify microsatellites in Funaria, and my class project with Sphagnum was going well and getting my advisor and other professors pretty excited. I, meanwhile, was feeling relieved to have real data to analyze without worrying about whether all the lab work would be successful. In the end, most of the major topics I am interested in will not change with Sphagnum: I can still study inbreeding, I can still study mating systems, and I have a better chance to study speciation than I did with Funaria. The reasons to switch kept piling up and I officially made up my mind on Friday morning.
Of course, because these things always happen that way, on Friday afternoon I received an e-mail. It was from Sigma Xi, the research organization, notifying me that I would be awarded a Grant in Research. The proposal I had written months ago concerned funding to develop microsatellites for Funaria. I now have the funding for the project I convinced myself I no longer want to do! Luckily, it shouldn't be a problem for Sigma Xi if I use the money to study Sphagnum instead. Still, the irony is palpable. In any event, I got my first graduate research grant!
Look forward in the coming weeks for a general Restart of Gametophyte Junction. Now that I'm more clear about which project I want to do, I can give lots of background and details!
Sunday, December 14, 2008
Thursday, June 26, 2008
Interlude: Moss Poetry
This just came in to my mailbox from BryoNet, the mailing list for the world's bryologists. It comes from Brent Mishler, who is a top bryologist at the University of California, Berkeley (formerly at Duke!). He sends along this poem on mosses, from nineteenth century Britishman William Gardiner. A clockmaker by trade, Gardiner was an avid collector of mosses and published a few of the first several important books on mosses. This poem appeared in one of them, Twenty Lessons on British Mosses, first published in 1846.
I think I can safely say this is the greatest moss poem ever written.
O! Let us love the silken moss
That clothes the time-worn wall
For great its Mighty Author is,
Although the plant be small.
The God who made the glorious sun
That shines so clear and bright,
And silver moon, and sparkling stars,
That gem the brow of night-
Did also give the sweet green moss
Its little form so fair;
And, though so tiny in all its parts,
Is not beneath His care.
When wandering in the fragrant wood,
Where pale primroses grow
To hear the tender ring-dove coo,
And happy small birds sing,
We tread a fresh and downy floor,
By soft green mosses made ;
And, when we rest by woodland stream,
Our couch with them is spread.
In valley deep, on mountain high-
The mosses still are there :
The dear delightful little things-
We meet them everywhere!
And when we mark them in our walks,
So beautiful, though small,
Our grateful hearts should glow with love
To Him who made them all.
I think I can safely say this is the greatest moss poem ever written.
Wednesday, June 18, 2008
Mr Johnson, I'm ready for my closeup...
Things have been fairly busy for me in the lab the last couple of weeks. I haven't been too successful with my molecular work. While that is pretty vital for my thesis, I've kept my spirits up by setting up a workflow for myself measuring phenotypes. My whole approach is aimed at integrating molecular data (genotype) with the size and shape of an organism (phenotype). Folks working on flowering plants have a lot more things to measure than I do, so I have to get the best out of my measurements. I'm intending on measuring seta size, capsule size, capsule shape, and spore number.
The last of these, spore number, is truly the most relevant to an evolutionary biologist. In terms of evolution, the most "fit" individuals are the ones who produce the most offspring. So by measuring the number of spores produced in a capsule, that's a pretty good idea of how many offspring that capsule is going to have. Why bother measuring anything else? Well, besides "my advisor says so," it may be that other characteristics about a sporophyte are more indicative of an inbred individual. For example, inbred Funaria may produce the same number of spores, but from a shorter seta- they may not be able to disperse as far, or they may get blocked out by taller sporophytes. Inbred capsules themselves may be smaller (even with the same number of spores), which may mean that per spore, an inbred capsule gets less nutrition.
I have been puzzling over how to measure such small organisms since I got back from my field trip. I met with various professors, including an expert on morphometrics (the science of measuring things). He suggested a program called ImageJ, software available for free and developed by the NIH. The software is pretty amazing. Here's my process:
1) Take a picture of the whole sporophyte, removed from the gametophyte. Here's an individual of Funaria flavicans, from Mechanicsville, NY:

2) Import the image into ImageJ. I have to calibrate the measurements using the milimeter scale in the bottom right, and then I seperate the image in to seta and capsule in 16-bit grayscale:

3) I "threshhold" the image, by telling the program what's an "object" and what's "not object" based on whether a particular pixel has any information in it. This effectively turns the image into a binary "object/not object" image:
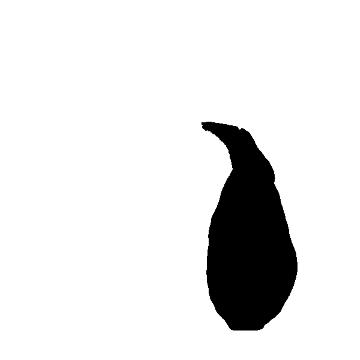
4) I use the "magic wand," familiar to anyone who's used a bit of image software before, to automatically draw a line hugging the outline of the binary image. I use the "Measure" command to automatically get the area inside the line, and the perimeter!
Taking the pictures using our microscope camera is the most time-consuming part. Once I have all the images it takes about half an hour to process twenty of them. The spore-counts will be a little different, and when I get started on them I'll tell you all how!
The last of these, spore number, is truly the most relevant to an evolutionary biologist. In terms of evolution, the most "fit" individuals are the ones who produce the most offspring. So by measuring the number of spores produced in a capsule, that's a pretty good idea of how many offspring that capsule is going to have. Why bother measuring anything else? Well, besides "my advisor says so," it may be that other characteristics about a sporophyte are more indicative of an inbred individual. For example, inbred Funaria may produce the same number of spores, but from a shorter seta- they may not be able to disperse as far, or they may get blocked out by taller sporophytes. Inbred capsules themselves may be smaller (even with the same number of spores), which may mean that per spore, an inbred capsule gets less nutrition.
I have been puzzling over how to measure such small organisms since I got back from my field trip. I met with various professors, including an expert on morphometrics (the science of measuring things). He suggested a program called ImageJ, software available for free and developed by the NIH. The software is pretty amazing. Here's my process:
1) Take a picture of the whole sporophyte, removed from the gametophyte. Here's an individual of Funaria flavicans, from Mechanicsville, NY:

2) Import the image into ImageJ. I have to calibrate the measurements using the milimeter scale in the bottom right, and then I seperate the image in to seta and capsule in 16-bit grayscale:

3) I "threshhold" the image, by telling the program what's an "object" and what's "not object" based on whether a particular pixel has any information in it. This effectively turns the image into a binary "object/not object" image:

4) I use the "magic wand," familiar to anyone who's used a bit of image software before, to automatically draw a line hugging the outline of the binary image. I use the "Measure" command to automatically get the area inside the line, and the perimeter!
Taking the pictures using our microscope camera is the most time-consuming part. Once I have all the images it takes about half an hour to process twenty of them. The spore-counts will be a little different, and when I get started on them I'll tell you all how!
Thursday, May 29, 2008
Nerd Beacon #2
Hey everyone. It's been a couple weeks since my last post, I've been hard at work trying to do multiple things to get my research going:
1. Find variable microsatellites so I can conduct population genetics studies.
2. Document my various collecting sites (also known as localities) using GPS and Google Earth, so I can have exact distances between collections.
3. Begin to measure traits (phenotypes) of the Funaria I collected, because the goal is to correlate relatedness with these traits.
I've been having trouble with all three, unfortunately. For the microsats, I've been having glitches with the equipment and my materials. One of our PCR machines broke while my experiment was running, and so that got ruined. I also am having trouble finding a program that lets me do all the sophisticated distance analysis I want that is also free. Finally, measuring traits is a bit difficult due to the shapes of the organisms.
In an evolutionary sense, the "fitness" of an organism is based upon how many offspring it has. For Funaria, the obvious thing to measure, then, is the number of spores each organism produces. However, this is a pretty time-consuming operation. Since each sporophyte produces 500,000 spores, I'm hoping I can use some other measure as a proxy for spore counts. One option is to measure the size of the capsule. However, this can be a pain because the capsule of Funaria is so oblique, rounded, and asymmetric. I'm meeting with a professor who specializes in measuring organisms tomorrow, so I hope he can help!
In the meantime, my only option is to measure the spores. The way I do this is by carefully rupturing a sporophyte into a small test tube that contains exactly one milliliter (mL) of sterile water. I then remove 1/10th of that (1 microliter) and place it on a special microscope slide known as a hemocytometer. The grids on that slide will allow me to count a small number of spores and then extrapolate to the whole sample. To count, I use what I consider to be nerd beacon #2- the clicker-counter:

This little device makes me happy on a number of levels- it keeps track of how many spores I see, sure. But more importantly, it is frequently used in baseball to keep track of pitch counts. Any combination of biology and baseball is definitely one I cherish.
1. Find variable microsatellites so I can conduct population genetics studies.
2. Document my various collecting sites (also known as localities) using GPS and Google Earth, so I can have exact distances between collections.
3. Begin to measure traits (phenotypes) of the Funaria I collected, because the goal is to correlate relatedness with these traits.
I've been having trouble with all three, unfortunately. For the microsats, I've been having glitches with the equipment and my materials. One of our PCR machines broke while my experiment was running, and so that got ruined. I also am having trouble finding a program that lets me do all the sophisticated distance analysis I want that is also free. Finally, measuring traits is a bit difficult due to the shapes of the organisms.
In an evolutionary sense, the "fitness" of an organism is based upon how many offspring it has. For Funaria, the obvious thing to measure, then, is the number of spores each organism produces. However, this is a pretty time-consuming operation. Since each sporophyte produces 500,000 spores, I'm hoping I can use some other measure as a proxy for spore counts. One option is to measure the size of the capsule. However, this can be a pain because the capsule of Funaria is so oblique, rounded, and asymmetric. I'm meeting with a professor who specializes in measuring organisms tomorrow, so I hope he can help!
In the meantime, my only option is to measure the spores. The way I do this is by carefully rupturing a sporophyte into a small test tube that contains exactly one milliliter (mL) of sterile water. I then remove 1/10th of that (1 microliter) and place it on a special microscope slide known as a hemocytometer. The grids on that slide will allow me to count a small number of spores and then extrapolate to the whole sample. To count, I use what I consider to be nerd beacon #2- the clicker-counter:
This little device makes me happy on a number of levels- it keeps track of how many spores I see, sure. But more importantly, it is frequently used in baseball to keep track of pitch counts. Any combination of biology and baseball is definitely one I cherish.
Thursday, May 15, 2008
Definition Train: Microsatellites
Next up stop for the definition train is not a concept but a technique, and one essential to my work. Microsatellites are also known as Short Tandem Repeats, and are DNA regions that contain a lot of repetitive DNA. Because they are very variable within a population, they're known as a "DNA Fingerprinting" technique, and are used for all kinds of purposes including forensics (they're featured on CSI quite often). Here's a little bit about how they are designed, and how they work.
Our DNA contains a lot of junk. That statement is less true than when it was first realized that despite the six billion base pairs of the human genome, less than a tenth of that is actually used for constructing proteins. Some of the rest of the genome is used to regulate gene expression, while much of it remains junk. However, junk is good for population geneticists, because junk DNA is not usually subject to natural selection; there aren't any constraints keeping a particular DNA sequence intact. They are free to vary according to the rules of chance, and those rules allow scientists to deduce how related two organisms are.
One type of these DNA regions contains repetitive bases- remember, the DNA code has four letters (A, G, C, and T). In these regions, the same two or three letters repeat many times. For example, it may look like this:
GATCAGTAAGATATATATATATATATATATATATATATATGGGTGCTCAGAT
In this case, this individual has the microsatellite "AT" repeated 16 times. The number of repeats turns out to be pretty variable, even within a population: one individual may have 16 repeats, as above, while a neighbor may have 12 repeats or 20 repeats. The interesting thing is that there are many of these regions (also known as loci) in the genome, and so one can have a pretty complete profile of an individual by looking at 5 to 15 of these loci:
Microsat Individual 1 Individual 2
1.............. 16................ 18
2............. 200 ............. 225
3.............. 75 ............... 75
4 ............. 87................ 90
5............ 103............... 115
This is a typical result of a microsat analysis; a machine reads off how many copies there are for each sample at each locus. With five loci, we can see that even though the two individuals are identical at locus 3, they are clearly two different individuals. Perhaps they are related, though. In forensics, crime labs will typically look at 13 loci to make a match between a suspect and a DNA sample from the crime scene. You might also hear on CSI or Court TV when it's said that the suspect has "7 of 13 alleles in common." What they mean is that at 7 of the 13 loci, two samples have identical numbers of repeats. This is extremely unlikely to be due to chance, and so it is pretty likely that the two samples come from DNA of blood-relatives (probably siblngs or parent-offspring).
It is one of my goals to investigate the matting patterns in mosses, by knowing who mates with whom. To do this, I need to find the regions (or loci) that have the microsatellites. Doing that from scratch is actually a time (3-4 months) and cost ($6000) intensive process. I'm fortunate that Funaria is in the same family as Physcomitrella, the lab rat of the moss world. Physcomitrella recently has its entire genome sequenced, and has a lot of researchers working on it, including those who have designed microsattelites for the species. In a paper, they also noted that many of the regions for Physcomitrella also work for Funaria. Those scientists have graciously supplied me with materials to discover whether those regions are variable enough for me to distinguish two moss individuals as they do with humans on CSI.
In addition, I hope to be able to tell whether the two species of Funaria (F. hygrometrica and F. flavicans) are hybridizing in nature. I would do this by examining populations where they are sympatric, and comparing them to populations which are allopatric. If the microsatellites show that the two species are more similar (by having more similar microsatellite profiles) when they are in sympatry than when in allopatry, it is likely that they are sharing genes.
Our DNA contains a lot of junk. That statement is less true than when it was first realized that despite the six billion base pairs of the human genome, less than a tenth of that is actually used for constructing proteins. Some of the rest of the genome is used to regulate gene expression, while much of it remains junk. However, junk is good for population geneticists, because junk DNA is not usually subject to natural selection; there aren't any constraints keeping a particular DNA sequence intact. They are free to vary according to the rules of chance, and those rules allow scientists to deduce how related two organisms are.
One type of these DNA regions contains repetitive bases- remember, the DNA code has four letters (A, G, C, and T). In these regions, the same two or three letters repeat many times. For example, it may look like this:
GATCAGTAAGATATATATATATATATATATATATATATATGGGTGCTCAGAT
In this case, this individual has the microsatellite "AT" repeated 16 times. The number of repeats turns out to be pretty variable, even within a population: one individual may have 16 repeats, as above, while a neighbor may have 12 repeats or 20 repeats. The interesting thing is that there are many of these regions (also known as loci) in the genome, and so one can have a pretty complete profile of an individual by looking at 5 to 15 of these loci:
Microsat Individual 1 Individual 2
1.............. 16................ 18
2............. 200 ............. 225
3.............. 75 ............... 75
4 ............. 87................ 90
5............ 103............... 115
This is a typical result of a microsat analysis; a machine reads off how many copies there are for each sample at each locus. With five loci, we can see that even though the two individuals are identical at locus 3, they are clearly two different individuals. Perhaps they are related, though. In forensics, crime labs will typically look at 13 loci to make a match between a suspect and a DNA sample from the crime scene. You might also hear on CSI or Court TV when it's said that the suspect has "7 of 13 alleles in common." What they mean is that at 7 of the 13 loci, two samples have identical numbers of repeats. This is extremely unlikely to be due to chance, and so it is pretty likely that the two samples come from DNA of blood-relatives (probably siblngs or parent-offspring).
It is one of my goals to investigate the matting patterns in mosses, by knowing who mates with whom. To do this, I need to find the regions (or loci) that have the microsatellites. Doing that from scratch is actually a time (3-4 months) and cost ($6000) intensive process. I'm fortunate that Funaria is in the same family as Physcomitrella, the lab rat of the moss world. Physcomitrella recently has its entire genome sequenced, and has a lot of researchers working on it, including those who have designed microsattelites for the species. In a paper, they also noted that many of the regions for Physcomitrella also work for Funaria. Those scientists have graciously supplied me with materials to discover whether those regions are variable enough for me to distinguish two moss individuals as they do with humans on CSI.
In addition, I hope to be able to tell whether the two species of Funaria (F. hygrometrica and F. flavicans) are hybridizing in nature. I would do this by examining populations where they are sympatric, and comparing them to populations which are allopatric. If the microsatellites show that the two species are more similar (by having more similar microsatellite profiles) when they are in sympatry than when in allopatry, it is likely that they are sharing genes.
Monday, May 12, 2008
Definition Train: Sympatry
It is my ongoing goal to explain, in common terms, what it is that I'm doing with this half-decade in graduate school. To that end, I'm hoping to make everyone familiar with certain terms; once you understand them, they're no longer over-your-head jargon! So every so often at the Gametophyte Junction we'll hop on the definition train. First up: sympatry.
Sympatry, like many biology terms, is a word derived from Greek: sym/syn- (meaning same), -patry (meaning fatherland). Two species are therefore sym-patric when they share the same fatherland; they have ranges which overlap. The opposite of sympatry is allopatry, meaning different-fatherland, and describes two species which have non-overlapping ranges. In between is parapatric, meaning adjacent-fatherland, and implying that the ranges of two species share a border (like two countries), or sometimes they have a narrow overlap range.
The concept of sympatry is important because of gene flow and speciation (the formation of new species). Initially, new species start out as populations, freely exchanging genes throughout the generations. The principles of population genetics have taught us that when a population has free gene flow, it is very tough to have different varieties. When a new variety emerges, three things usually happen: 1) The new variety is more adapted to the environment, and spreads quickly, wiping out the older varieties. 2) The new variety is less adapted to the environment, and vanishes quickly. 3) The new variety is neutral with respect to how adapted to the environment it is, and these varieties fluctuate until they are eventually lost or adopted by the whole population.
Species, meanwhile, under the Biological Species Concept, are not just observed by their separate and discontinuous variety. To achieve this discontinuity, there must be some barrier to gene flow between the population with the new variety and the other populations. Typically, this is done in allopatry- some geographic barrier (like a mountain or a body of water) separates two populations for an extended period of time. During this time, the two populations adapt to environments that are almost certainly different. These different adaptations may also change the organism in such a way that if the barrier were removed (a gap in the mountain, land bridge over the body of water), the two populations may have trouble mating. This creates the barrier to gene flow.
You'll notice that when the barrier is removed, the two populations are now in sympatry. It is usually at this point that species are truly formed- when two populations have reduced gene flow among them in sympatry. On a practical level, this manifests usually as "hybrid breakdown." Most people are familiar with the mule- it is an offspring of a horse and a donkey, but the mule itself is sterile. Because the horse and donkey adapted to different environments, they evolved genes that are slightly incompatible; enough to create a live child but one that is sterile. Having sterile offspring is definitely a barrier to gene flow. It is also a waste of resources, and natural selection typically leads such populations to methods of avoiding creating these bad hybrids. Animals recognize members of their own species through sight and sound; plants change pollinators or chemicals on the surfaces of gametes in an effort to only use precious reproduction resources on making good children.
What does this have to do with me? Well, by and large most of the work on mosses has not been under the Biological Species Concept. Bryologists over the years have been guided by the principles of taxonomy, grouping organisms in to species and genera and families based on similar characteristics. Whether two moss species have any barriers to gene flow is largely unknown. So it is entirely possible that the two species I'm studying, Funaria hygrometrica and Funaria flavicans, actually exchange genes. If so, then they are, in my opinion, one species. I'm bringing this up in part because of the following picture:

This was from my field trip, taken near Dillwyn, Virginia. The green capsules are F. hygrometrica and the smaller red capsules are F. flavicans. They are growing right next to each other, sympatrically. This is not a unique finding, my advisor wrote a paper in 1992 describing a mine site which had both species and what appeared to be intermediates. However, no DNA work was done to determine if there were hybrid plants. Some of the questions that intrigue me include:
Do the two species hybridize? If not, what barrier to gene flow separates them?
If hybrids form, are they less "fit" than their parents?
What genes control the separation of the species? Are there many genes or just a few?
For example, the F. flavicans in the picture above are red because they matured earlier than F. hygrometrica. Timing in plant matings are very important; if hybrids between the species were to mature at some intermediate time, they may not find many mates. That would make them less "fit" because "number of offspring" is the definition of fitness.
That's all for now; as always, feel free to ask questions if there's something unclear. I'm always willing to find new ways to explain things!
Sympatry, like many biology terms, is a word derived from Greek: sym/syn- (meaning same), -patry (meaning fatherland). Two species are therefore sym-patric when they share the same fatherland; they have ranges which overlap. The opposite of sympatry is allopatry, meaning different-fatherland, and describes two species which have non-overlapping ranges. In between is parapatric, meaning adjacent-fatherland, and implying that the ranges of two species share a border (like two countries), or sometimes they have a narrow overlap range.
The concept of sympatry is important because of gene flow and speciation (the formation of new species). Initially, new species start out as populations, freely exchanging genes throughout the generations. The principles of population genetics have taught us that when a population has free gene flow, it is very tough to have different varieties. When a new variety emerges, three things usually happen: 1) The new variety is more adapted to the environment, and spreads quickly, wiping out the older varieties. 2) The new variety is less adapted to the environment, and vanishes quickly. 3) The new variety is neutral with respect to how adapted to the environment it is, and these varieties fluctuate until they are eventually lost or adopted by the whole population.
Species, meanwhile, under the Biological Species Concept, are not just observed by their separate and discontinuous variety. To achieve this discontinuity, there must be some barrier to gene flow between the population with the new variety and the other populations. Typically, this is done in allopatry- some geographic barrier (like a mountain or a body of water) separates two populations for an extended period of time. During this time, the two populations adapt to environments that are almost certainly different. These different adaptations may also change the organism in such a way that if the barrier were removed (a gap in the mountain, land bridge over the body of water), the two populations may have trouble mating. This creates the barrier to gene flow.
You'll notice that when the barrier is removed, the two populations are now in sympatry. It is usually at this point that species are truly formed- when two populations have reduced gene flow among them in sympatry. On a practical level, this manifests usually as "hybrid breakdown." Most people are familiar with the mule- it is an offspring of a horse and a donkey, but the mule itself is sterile. Because the horse and donkey adapted to different environments, they evolved genes that are slightly incompatible; enough to create a live child but one that is sterile. Having sterile offspring is definitely a barrier to gene flow. It is also a waste of resources, and natural selection typically leads such populations to methods of avoiding creating these bad hybrids. Animals recognize members of their own species through sight and sound; plants change pollinators or chemicals on the surfaces of gametes in an effort to only use precious reproduction resources on making good children.
What does this have to do with me? Well, by and large most of the work on mosses has not been under the Biological Species Concept. Bryologists over the years have been guided by the principles of taxonomy, grouping organisms in to species and genera and families based on similar characteristics. Whether two moss species have any barriers to gene flow is largely unknown. So it is entirely possible that the two species I'm studying, Funaria hygrometrica and Funaria flavicans, actually exchange genes. If so, then they are, in my opinion, one species. I'm bringing this up in part because of the following picture:

This was from my field trip, taken near Dillwyn, Virginia. The green capsules are F. hygrometrica and the smaller red capsules are F. flavicans. They are growing right next to each other, sympatrically. This is not a unique finding, my advisor wrote a paper in 1992 describing a mine site which had both species and what appeared to be intermediates. However, no DNA work was done to determine if there were hybrid plants. Some of the questions that intrigue me include:
Do the two species hybridize? If not, what barrier to gene flow separates them?
If hybrids form, are they less "fit" than their parents?
What genes control the separation of the species? Are there many genes or just a few?
For example, the F. flavicans in the picture above are red because they matured earlier than F. hygrometrica. Timing in plant matings are very important; if hybrids between the species were to mature at some intermediate time, they may not find many mates. That would make them less "fit" because "number of offspring" is the definition of fitness.
That's all for now; as always, feel free to ask questions if there's something unclear. I'm always willing to find new ways to explain things!
Thursday, May 8, 2008
Days 6&7&9: Homeward Bound (with Pictures!)
So, it's been almost a week since my last post. I apologize for that, it has been a long busy week, and I've been splitting my time between the greenhouse and helping Rebecca find an apartment in Durham (she's working at the Duke Lemur Center next year!) I'd like to complete the series about my trip. I've also gotten my pictures uploaded, so I can finally share with you all some of the places I've been. Click HERE for the whole Google album!!!
Image embedding seems to not work very well, so I'm just going to link to pictures. Click on them for a thousand extra words. For starters, here's Funaria in all its glory.That's from Tompkins, County New York, near West Danby, in the really interesting population I talked about here.
We were able to collect two populations on Friday, one in Charlestown, Rhode Island and another in Plainfield, Connecticut. Oddly enough, they were both along railroad tracks parallel to side streets each named "Railroad Avenue." We didn't do too much else on Friday, although we did collect mosses from around Rhode Island and visited Beavertail Lighthouse, at the southern tip of Conanicut Island (in the sound between Providence and Newport). It's the third-oldest lighthouse in the US, according to the signs. Here's me at the lighthouse, trying not to be cold.
After making it home on Friday night we enjoyed an excellent meal of Carolina-style BBQ. Piers seemed to enjoy it despite his preference for Memphis-style, and I've been eating the leftovers back in Durham all week. On Saturday morning, Piers and I got up and traveled to the exotic location of... about a mile from our house, in the Wallkill River Wildlife Refuge. The Duke Herbarium has only six collections from Sussex, County NJ, and most of the collections in the New York Botanical Garden are pre-1950 or even pre-1900. So everything we collected was useful for bryologists in general. We went over to Vernon to the railroad tracks and found a quite extensive population of Funaria there.
There was also a very nice older man from the shops across the street, Dan Børstad. At first
he thought we were trash collectors, but after I explained what we were doing, he was still happy to talk to me. He was from Norway and said he had studied Botany when there, and I told him about all of our Norwegian colleagues who study Sphagnum up in the massive peat bogs. He also seemed impressed that I had returned all the way home just to collect the moss.
In the afternoon, Dad wanted to join us to see how the whole moss collecting operates. We were mostly searching for Sphagnum, which is the only moss that has been collected from the county. Most of the habitats we were looking at, however, looked more like this, which is far too eutrophic for Sphagnum.
We found a decent sized Funaria population (where I put Dad to work) in Sparta and hunted around for some of the previous localities for Sphagnum. One place we visited was Edison Pond, which I did not know existed. It's about 2 miles SSE of Ogdensburg, and was a series of mines funded by Thomas Edison in the early 1900s. He poured $2 million of his own money into the site to mine for iron, and employed 500 men at the peak of construction. There was no iron there, however, as the Hamburg Mountains are made almost entirely out of zinc and lime. The pond, and presumably the Sphagnum collected in 1986, were also gone.
Leaving Edison Pond, we headed up to High Point, and passed on entering the park itself to visit a swampy area at the base of Steenykill Lake, and found copious amounts of Sphagnum! Sadly, there weren't any sporophytes, but I'm hoping to come home again sometime in July, and perhaps I will be luckier then. The grad student working on them, however, is less convinced, and she thinks these species of Sphagnum never enter the sexual stage. How sad.
We settled in for an evening of Lasagana and a moss show-and-tell for my parents. The next morning we left, laden with moss and bagels from New Jersey, headed back to Durham. We stopped back in Bethesda for lunch, and then once in Catlett, Virginia to make a final Funaria collection.
It's going to be a lengthy job to sort out and document all the Funaria I collected during this trip. I had a lot of fun though, learning not just about the species I'm focusing on, but also gaining more field experience with mosses by traveling with such a knowledgeable guide. Even five days after returning, I'm thinking back upon what a whirlwind trip it was. I've already discovered some very intruiging things about the plants I collected this week; I hope to share some of those discoveries with you soon. So, by all means do not think that the end of my trip signals the end of my blogging- there's many more Fun(aria) to come!
Total Miles Traveled: 1,987
Total Funaria Populations: 18
Oil Pans Replaced: 1
Clichéd Advertising Reference: Priceless.
Image embedding seems to not work very well, so I'm just going to link to pictures. Click on them for a thousand extra words. For starters, here's Funaria in all its glory.That's from Tompkins, County New York, near West Danby, in the really interesting population I talked about here.
We were able to collect two populations on Friday, one in Charlestown, Rhode Island and another in Plainfield, Connecticut. Oddly enough, they were both along railroad tracks parallel to side streets each named "Railroad Avenue." We didn't do too much else on Friday, although we did collect mosses from around Rhode Island and visited Beavertail Lighthouse, at the southern tip of Conanicut Island (in the sound between Providence and Newport). It's the third-oldest lighthouse in the US, according to the signs. Here's me at the lighthouse, trying not to be cold.
After making it home on Friday night we enjoyed an excellent meal of Carolina-style BBQ. Piers seemed to enjoy it despite his preference for Memphis-style, and I've been eating the leftovers back in Durham all week. On Saturday morning, Piers and I got up and traveled to the exotic location of... about a mile from our house, in the Wallkill River Wildlife Refuge. The Duke Herbarium has only six collections from Sussex, County NJ, and most of the collections in the New York Botanical Garden are pre-1950 or even pre-1900. So everything we collected was useful for bryologists in general. We went over to Vernon to the railroad tracks and found a quite extensive population of Funaria there.
There was also a very nice older man from the shops across the street, Dan Børstad. At first
he thought we were trash collectors, but after I explained what we were doing, he was still happy to talk to me. He was from Norway and said he had studied Botany when there, and I told him about all of our Norwegian colleagues who study Sphagnum up in the massive peat bogs. He also seemed impressed that I had returned all the way home just to collect the moss.
In the afternoon, Dad wanted to join us to see how the whole moss collecting operates. We were mostly searching for Sphagnum, which is the only moss that has been collected from the county. Most of the habitats we were looking at, however, looked more like this, which is far too eutrophic for Sphagnum.
We found a decent sized Funaria population (where I put Dad to work) in Sparta and hunted around for some of the previous localities for Sphagnum. One place we visited was Edison Pond, which I did not know existed. It's about 2 miles SSE of Ogdensburg, and was a series of mines funded by Thomas Edison in the early 1900s. He poured $2 million of his own money into the site to mine for iron, and employed 500 men at the peak of construction. There was no iron there, however, as the Hamburg Mountains are made almost entirely out of zinc and lime. The pond, and presumably the Sphagnum collected in 1986, were also gone.
Leaving Edison Pond, we headed up to High Point, and passed on entering the park itself to visit a swampy area at the base of Steenykill Lake, and found copious amounts of Sphagnum! Sadly, there weren't any sporophytes, but I'm hoping to come home again sometime in July, and perhaps I will be luckier then. The grad student working on them, however, is less convinced, and she thinks these species of Sphagnum never enter the sexual stage. How sad.
We settled in for an evening of Lasagana and a moss show-and-tell for my parents. The next morning we left, laden with moss and bagels from New Jersey, headed back to Durham. We stopped back in Bethesda for lunch, and then once in Catlett, Virginia to make a final Funaria collection.
It's going to be a lengthy job to sort out and document all the Funaria I collected during this trip. I had a lot of fun though, learning not just about the species I'm focusing on, but also gaining more field experience with mosses by traveling with such a knowledgeable guide. Even five days after returning, I'm thinking back upon what a whirlwind trip it was. I've already discovered some very intruiging things about the plants I collected this week; I hope to share some of those discoveries with you soon. So, by all means do not think that the end of my trip signals the end of my blogging- there's many more Fun(aria) to come!
Total Miles Traveled: 1,987
Total Funaria Populations: 18
Oil Pans Replaced: 1
Clichéd Advertising Reference: Priceless.
Subscribe to:
Comments (Atom)
