Things have been fairly busy for me in the lab the last couple of weeks. I haven't been too successful with my molecular work. While that is pretty vital for my thesis, I've kept my spirits up by setting up a workflow for myself measuring phenotypes. My whole approach is aimed at integrating molecular data (genotype) with the size and shape of an organism (phenotype). Folks working on flowering plants have a lot more things to measure than I do, so I have to get the best out of my measurements. I'm intending on measuring seta size, capsule size, capsule shape, and spore number.
The last of these, spore number, is truly the most relevant to an evolutionary biologist. In terms of evolution, the most "fit" individuals are the ones who produce the most offspring. So by measuring the number of spores produced in a capsule, that's a pretty good idea of how many offspring that capsule is going to have. Why bother measuring anything else? Well, besides "my advisor says so," it may be that other characteristics about a sporophyte are more indicative of an inbred individual. For example, inbred Funaria may produce the same number of spores, but from a shorter seta- they may not be able to disperse as far, or they may get blocked out by taller sporophytes. Inbred capsules themselves may be smaller (even with the same number of spores), which may mean that per spore, an inbred capsule gets less nutrition.
I have been puzzling over how to measure such small organisms since I got back from my field trip. I met with various professors, including an expert on morphometrics (the science of measuring things). He suggested a program called ImageJ, software available for free and developed by the NIH. The software is pretty amazing. Here's my process:
1) Take a picture of the whole sporophyte, removed from the gametophyte. Here's an individual of Funaria flavicans, from Mechanicsville, NY:

2) Import the image into ImageJ. I have to calibrate the measurements using the milimeter scale in the bottom right, and then I seperate the image in to seta and capsule in 16-bit grayscale:

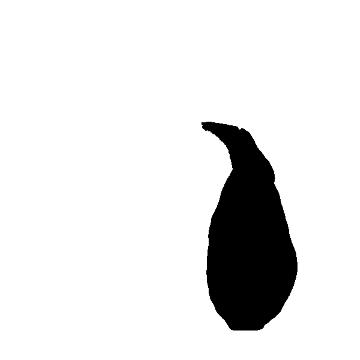
3) I "threshhold" the image, by telling the program what's an "object" and what's "not object" based on whether a particular pixel has any information in it. This effectively turns the image into a binary "object/not object" image:
4) I use the "magic wand," familiar to anyone who's used a bit of image software before, to automatically draw a line hugging the outline of the binary image. I use the "Measure" command to automatically get the area inside the line, and the perimeter!
Taking the pictures using our microscope camera is the most time-consuming part. Once I have all the images it takes about half an hour to process twenty of them. The spore-counts will be a little different, and when I get started on them I'll tell you all how!
The last of these, spore number, is truly the most relevant to an evolutionary biologist. In terms of evolution, the most "fit" individuals are the ones who produce the most offspring. So by measuring the number of spores produced in a capsule, that's a pretty good idea of how many offspring that capsule is going to have. Why bother measuring anything else? Well, besides "my advisor says so," it may be that other characteristics about a sporophyte are more indicative of an inbred individual. For example, inbred Funaria may produce the same number of spores, but from a shorter seta- they may not be able to disperse as far, or they may get blocked out by taller sporophytes. Inbred capsules themselves may be smaller (even with the same number of spores), which may mean that per spore, an inbred capsule gets less nutrition.
I have been puzzling over how to measure such small organisms since I got back from my field trip. I met with various professors, including an expert on morphometrics (the science of measuring things). He suggested a program called ImageJ, software available for free and developed by the NIH. The software is pretty amazing. Here's my process:
1) Take a picture of the whole sporophyte, removed from the gametophyte. Here's an individual of Funaria flavicans, from Mechanicsville, NY:

2) Import the image into ImageJ. I have to calibrate the measurements using the milimeter scale in the bottom right, and then I seperate the image in to seta and capsule in 16-bit grayscale:

3) I "threshhold" the image, by telling the program what's an "object" and what's "not object" based on whether a particular pixel has any information in it. This effectively turns the image into a binary "object/not object" image:

4) I use the "magic wand," familiar to anyone who's used a bit of image software before, to automatically draw a line hugging the outline of the binary image. I use the "Measure" command to automatically get the area inside the line, and the perimeter!
Taking the pictures using our microscope camera is the most time-consuming part. Once I have all the images it takes about half an hour to process twenty of them. The spore-counts will be a little different, and when I get started on them I'll tell you all how!

No comments:
Post a Comment